
The aim of the prcbench package is to provide a testing workbench for evaluating precision-recall curves under various conditions. It contains integrated interfaces for the following five tools. It also contains predefined test data sets.
| Tool | Link |
|---|---|
| ROCR | Tool web site, CRAN |
| AUCCalculator | Tool web site |
| PerfMeas | CRAN |
| PRROC | CRAN |
| precrec | Tool web site, CRAN |
Introduction to prcbench – a package vignette that contains the descriptions of the functions with several useful examples. View the vignette with vignette("introduction", package = "prcbench") in R. The HTML version is also available on the GitHub Pages.
Help pages – all the functions including the S3 generics have their own help pages with plenty of examples. View the main help page with help(package = "prcbench") in R. The HTML version is also available on the GitHub Pages.
AUCCalculator requires a Java runtime (>= 6).
PerfMeas requires Bioconductor libraries.
Install the release version of prcbench from CRAN with install.packages("prcbench").
Alternatively, you can install a development version of prcbench from our GitHub repository. To install it:
Make sure you have a working development environment.
Install devtools from CRAN with install.packages("devtools").
Install prcbench from the GitHub repository with devtools::install_github("evalclass/prcbench").
You can manually install the dependencies from Bioconductor if install.packages fails to access the Bioconductor repository.
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("limma")
BiocManager::install("graph")
BiocManager::install("RBGL")microbenchmark does not work on some OSs. prcbench uses system.time when microbenchmark is not available.
sudo R CMD javareconfFollowing two examples show the basic usage of prcbench functions.
The run_benchmark function outputs the result of microbenchmark for specified tools.
## Load library
library(prcbench)
## Run microbenchmark for auc5 (five tools) on b10 (balanced 5 Ps and 5 Ns)
testset <- create_testset("bench", "b10")
toolset <- create_toolset(set_names = "auc5")
res <- run_benchmark(testset, toolset)
## Use knitr::kable to show the result in a table format
knitr::kable(res$tab, digits = 2)| testset | toolset | toolname | min | lq | mean | median | uq | max | neval |
|---|---|---|---|---|---|---|---|---|---|
| b10 | auc5 | AUCCalculator | 3.00 | 3.98 | 8.07 | 4.90 | 5.45 | 23.04 | 5 |
| b10 | auc5 | PerfMeas | 0.06 | 0.07 | 85.07 | 0.07 | 0.10 | 425.04 | 5 |
| b10 | auc5 | precrec | 5.56 | 6.13 | 42.58 | 7.43 | 12.04 | 181.75 | 5 |
| b10 | auc5 | PRROC | 0.18 | 0.19 | 1.82 | 0.19 | 0.23 | 8.32 | 5 |
| b10 | auc5 | ROCR | 1.89 | 1.99 | 12.33 | 2.13 | 17.08 | 38.56 | 5 |
The run_evalcurve function evaluates precision-recall curves with predefined test datasets. The autoplot shows a plot with the result of the run_evalcurve function.
## ggplot2 is necessary to use autoplot
library(ggplot2)
## Plot base points and the result of precrec on c1, c2, and c3 test sets
testset <- create_testset("curve", c("c1", "c2", "c3"))
toolset <- create_toolset("precrec")
scores1 <- run_evalcurve(testset, toolset)
autoplot(scores1)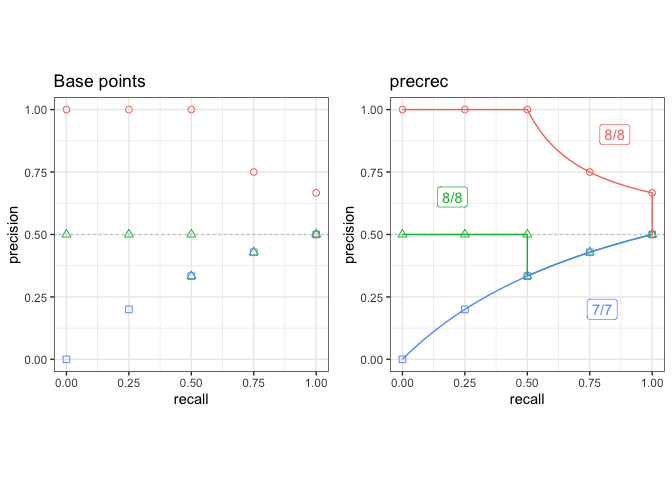
## Plot the results of PerfMeas and PRROC on c1, c2, and c3 test sets
toolset <- create_toolset(c("PerfMeas", "PRROC"))
scores2 <- run_evalcurve(testset, toolset)
autoplot(scores2, base_plot = FALSE)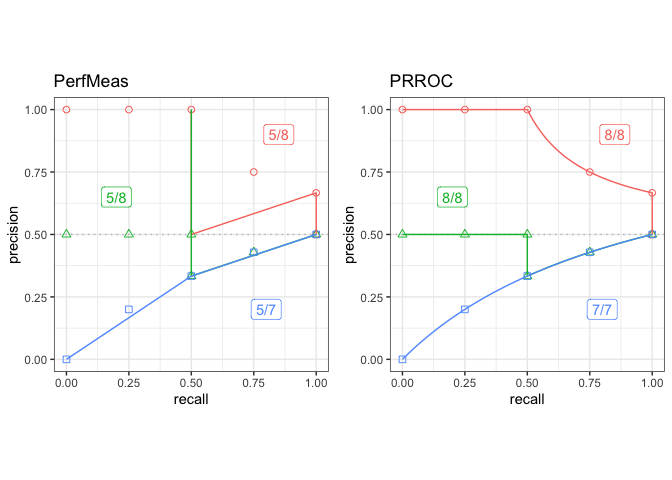
Precrec: fast and accurate precision-recall and ROC curve calculations in R
Takaya Saito; Marc Rehmsmeier
Bioinformatics 2017; 33 (1): 145-147.
doi: 10.1093/bioinformatics/btw570
Classifier evaluation with imbalanced datasets – our web site that contains several pages with useful tips for performance evaluation on binary classifiers.
The Precision-Recall Plot Is More Informative than the ROC Plot When Evaluating Binary Classifiers on Imbalanced Datasets – our paper that summarized potential pitfalls of ROC plots with imbalanced datasets and advantages of using precision-recall plots instead.