The covid19srilanka package provides a tidy format dataset of the 2019 Novel Coronavirus COVID-19 (2019-nCoV) epidemic in Sri Lanka
You can install the development version from GitHub with:
data("covid.cases")
head(covid.cases)
#> Date Type Count
#> 1 2020-03-29 Confirmed 115
#> 2 2020-03-29 Recovered 10
#> 3 2020-03-29 Deaths 1
#> 4 2020-03-29 Active 104
#> 5 2020-03-30 Confirmed 120
#> 6 2020-03-30 Recovered 11
tail(covid.cases)
#> Date Type Count
#> 2195 2021-09-29 Deaths 12786
#> 2196 2021-09-29 Actives 45881
#> 2197 2021-09-30 Confirmed 516465
#> 2198 2021-09-30 Recovered 457488
#> 2199 2021-09-30 Deaths 12847
#> 2200 2021-09-30 Actives 46130data("district.wise.cases")
head(district.wise.cases)
#> Date District Count
#> 1 2021-08-01 Colombo 71267
#> 2 2021-08-01 Gampaha 56085
#> 3 2021-08-01 Kalutara 33300
#> 4 2021-08-01 Kandy 14576
#> 5 2021-08-01 Kurunagala 15327
#> 6 2021-08-01 Galle 14841data("vaccination")
head(vaccination)
#> Date Vaccine first dose Second dose
#> 1 2021-04-29 Covishield Vaccine 925242 3445
#> 2 2021-04-29 Sinopharm Vaccine 2469 2435
#> 3 2021-04-30 Covishield Vaccine 925242 22919
#> 4 2021-04-30 Sinopharm Vaccine 2865 2435
#> 5 2021-05-01 Covishield Vaccine 925242 60757
#> 6 2021-05-01 Sinopharm Vaccine 2865 2435library(tidyverse)
#> ── Attaching packages ─────────────────────────────────────── tidyverse 1.3.1 ──
#> ✓ ggplot2 3.3.5 ✓ purrr 0.3.4
#> ✓ tibble 3.1.5 ✓ dplyr 1.0.7
#> ✓ tidyr 1.1.4 ✓ stringr 1.4.0
#> ✓ readr 2.0.2 ✓ forcats 0.5.1
#> ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
#> x dplyr::filter() masks stats::filter()
#> x dplyr::lag() masks stats::lag()
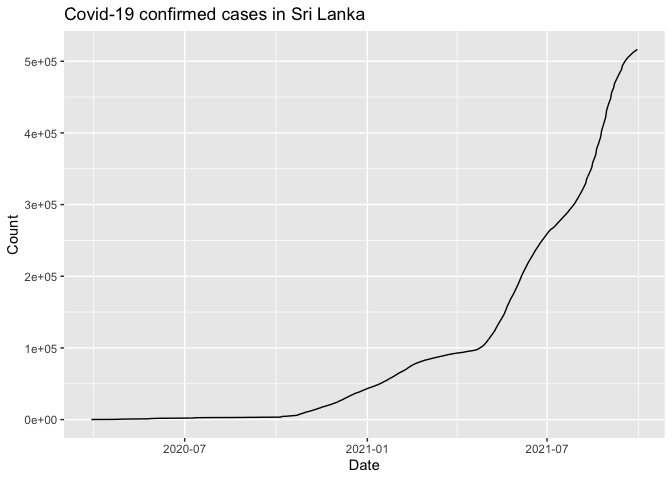
covid.cases %>% filter(Type=="Confirmed") %>% ggplot(aes(x=Date, y=Count)) + geom_line() + ggtitle("Covid-19 confirmed cases in Sri Lanka")