The goal of FFTrees is to create and visualize fast-and-frugal decision trees (FFTs) from data with a binary outcome following the methods described in Phillips, Neth, Woike & Gaissmaier (2017).
You can install the released version of FFTrees from CRAN with:
install.packages("FFTrees")And the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("ndphillips/FFTrees", build_vignettes = TRUE)library(FFTrees)
#>
#> O
#> / \
#> F O
#> / \
#> F Trees 1.6.6
#>
#> Email: Nathaniel.D.Phillips.is@gmail.com
#> FFTrees.guide() opens the main guide.Let’s create a fast-and-frugal tree predicting heart disease status
(“Healthy” vs. “Diseased”) based on a heart.train dataset,
and test it on heart.test a testing dataset.
Here are the first new rows and columns of our datasets. The key
column is diagnosis, a logical column (TRUE and FALSE)
which indicate, for each patient, whether or not they have heart
disease.
Here is heart.train (the training dataset) which
contains data from 150 patients:
heart.train
#> # A tibble: 150 × 14
#> diagnosis age sex cp trestbps chol fbs restecg thalach exang
#> <lgl> <dbl> <dbl> <chr> <dbl> <dbl> <dbl> <chr> <dbl> <dbl>
#> 1 FALSE 44 0 np 108 141 0 normal 175 0
#> 2 FALSE 51 0 np 140 308 0 hypertrophy 142 0
#> 3 FALSE 52 1 np 138 223 0 normal 169 0
#> 4 TRUE 48 1 aa 110 229 0 normal 168 0
#> 5 FALSE 59 1 aa 140 221 0 normal 164 1
#> 6 FALSE 58 1 np 105 240 0 hypertrophy 154 1
#> 7 FALSE 41 0 aa 126 306 0 normal 163 0
#> 8 TRUE 39 1 a 118 219 0 normal 140 0
#> 9 TRUE 77 1 a 125 304 0 hypertrophy 162 1
#> 10 FALSE 41 0 aa 105 198 0 normal 168 0
#> # … with 140 more rows, and 4 more variables: oldpeak <dbl>, slope <chr>,
#> # ca <dbl>, thal <chr>Here is heart.test (the testing / prediction dataset)
which contains data from a new set of 153 patients:
heart.test
#> # A tibble: 153 × 14
#> diagnosis age sex cp trestbps chol fbs restecg thalach exang
#> <lgl> <dbl> <dbl> <chr> <dbl> <dbl> <dbl> <chr> <dbl> <dbl>
#> 1 FALSE 51 0 np 120 295 0 hypertrophy 157 0
#> 2 TRUE 45 1 ta 110 264 0 normal 132 0
#> 3 TRUE 53 1 a 123 282 0 normal 95 1
#> 4 TRUE 45 1 a 142 309 0 hypertrophy 147 1
#> 5 FALSE 66 1 a 120 302 0 hypertrophy 151 0
#> 6 TRUE 48 1 a 130 256 1 hypertrophy 150 1
#> 7 TRUE 55 1 a 140 217 0 normal 111 1
#> 8 FALSE 56 1 aa 130 221 0 hypertrophy 163 0
#> 9 TRUE 42 1 a 136 315 0 normal 125 1
#> 10 FALSE 45 1 a 115 260 0 hypertrophy 185 0
#> # … with 143 more rows, and 4 more variables: oldpeak <dbl>, slope <chr>,
#> # ca <dbl>, thal <chr>Now let’s use FFTrees() to create a fast and frugal tree
from the heart.train data and test their performance on
heart.test
# Create an FFTrees object from the heartdisease data
heart.fft <- FFTrees(formula = diagnosis ~.,
data = heart.train,
data.test = heart.test,
decision.labels = c("Healthy", "Disease"))
#> Setting goal = 'wacc'
#> Setting goal.chase = 'waccc'
#> Setting cost.outcomes = list(hi = 0, mi = 1, fa = 1, cr = 0)
#> Growing FFTs with ifan
#> Fitting other algorithms for comparison (disable with do.comp = FALSE) ...
# See the print method which shows aggregatge statistics
heart.fft
#> FFTrees
#> - Trees: 7 fast-and-frugal trees predicting diagnosis
#> - Outcome costs: [hi = 0, mi = 1, fa = 1, cr = 0]
#>
#> FFT #1: Definition
#> [1] If thal = {rd,fd}, decide Disease.
#> [2] If cp != {a}, decide Healthy.
#> [3] If ca <= 0, decide Healthy, otherwise, decide Disease.
#>
#> FFT #1: Prediction Accuracy
#> Prediction Data: N = 153, Pos (+) = 73 (48%)
#>
#> | | True + | True - |
#> |---------|--------|--------|
#> |Decide + | hi 64 | fa 19 | 83
#> |Decide - | mi 9 | cr 61 | 70
#> |---------|--------|--------|
#> 73 80 N = 153
#>
#> acc = 81.7% ppv = 77.1% npv = 87.1%
#> bacc = 82.0% sens = 87.7% spec = 76.2%
#> E(cost) = 0.183
#>
#> FFT #1: Prediction Speed and Frugality
#> mcu = 1.73, pci = 0.87
# Plot the best tree applied to the test data
plot(heart.fft,
data = "test",
main = "Heart Disease")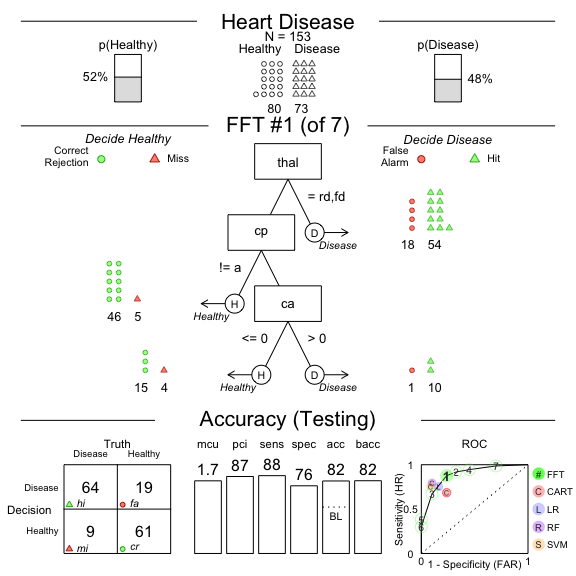
# Compare results across algorithms in test data
heart.fft$competition$test
#> algorithm n hi fa mi cr sens spec far ppv npv
#> 1 fftrees 153 64 19 9 61 0.8767123 0.7625 0.2375 0.7710843 0.8714286
#> 2 lr 153 55 13 18 67 0.7534247 0.8375 0.1625 0.8088235 0.7882353
#> 3 cart 153 50 19 23 61 0.6849315 0.7625 0.2375 0.7246377 0.7261905
#> 4 rf 153 58 8 15 72 0.7945205 0.9000 0.1000 0.8787879 0.8275862
#> 5 svm 153 55 7 18 73 0.7534247 0.9125 0.0875 0.8870968 0.8021978
#> acc bacc cost cost_decisions cost_cues
#> 1 0.8169935 0.8196062 0.1830065 0.1830065 0
#> 2 0.7973856 0.7954623 0.2026144 0.2026144 NA
#> 3 0.7254902 0.7237158 0.2745098 0.2745098 NA
#> 4 0.8496732 0.8472603 0.1503268 0.1503268 NA
#> 5 0.8366013 0.8329623 0.1633987 0.1633987 NABecause fast-and-frugal trees are so simple, you can create one ‘from words’ and apply it to data!
For example, below we’ll create a tree with the following 4 nodes and
evaluate its performance on the heart.test data
# Create your own custom FFT 'in words' and apply it to data
# Create my own fft
my.fft <- FFTrees(formula = diagnosis ~.,
data = heart.train,
data.test = heart.test,
decision.labels = c("Healthy", "Disease"),
my.tree = "If sex = 1, predict Disease.
If age < 45, predict Healthy.
If thal = {fd, normal}, predict Disease.
Otherwise, predict Healthy")
#> Setting goal = 'wacc'
#> Setting goal.chase = 'waccc'
#> Setting cost.outcomes = list(hi = 0, mi = 1, fa = 1, cr = 0)
#> Fitting other algorithms for comparison (disable with do.comp = FALSE) ...
# Plot my custom fft and see how it did
plot(my.fft,
data = "test",
main = "Custom FFT")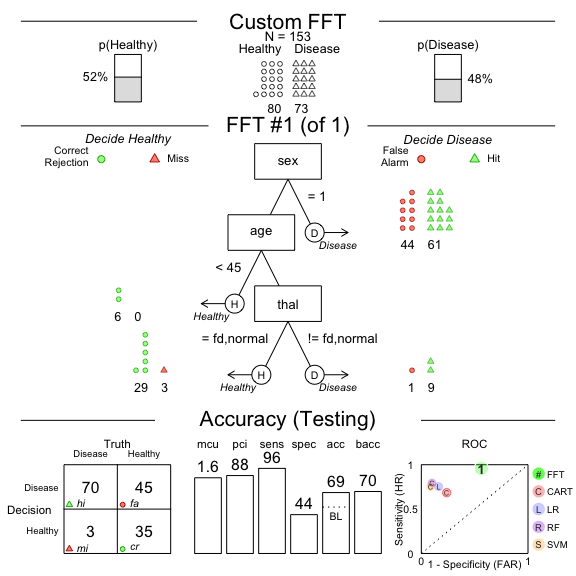
As we can see, the tree has very high sensitivity but terrible specificity.
APA Citation
Phillips, Nathaniel D., Neth, Hansjoerg, Woike, Jan K., & Gaissmaier, W. (2017). FFTrees: A toolbox to create, visualize, and evaluate fast-and-frugal decision trees. Judgment and Decision Making, 12(4), 344-368.
We had a lot of fun creating FFTrees and hope you like it too! We
have an article introducing the FFTrees package in the
journal Judgment and Decision Making titled FFTrees: A toolbox to
create, visualize,and evaluate fast-and-frugal decision trees. We
encourage you to read the article to learn more about the history of
FFTs and how the FFTrees package creates them.
If you use FFTrees in your work, please cite us and spread the word so we can continue developing the package
Here are some example publications that have used FFTrees (find the full list at Google Scholar)